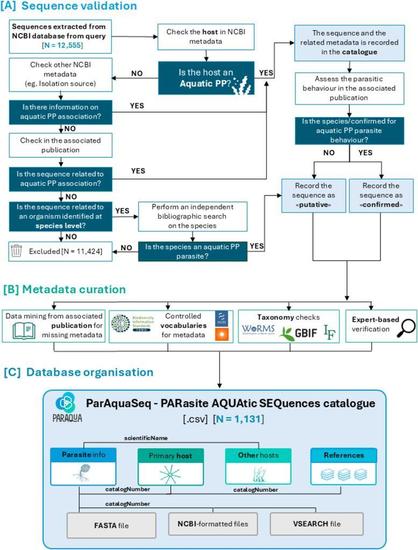
ParAquaSeq, a Database of Ecologically Annotated rRNA Sequences Covering Zoosporic Parasites Infecting Aquatic Primary Producers in Natural and Industrial Systems #eDNA #metabarcoding #protists https://onlinelibrary.wiley.com/doi/10.1111/1755-0998.14099
#Metabarcoding
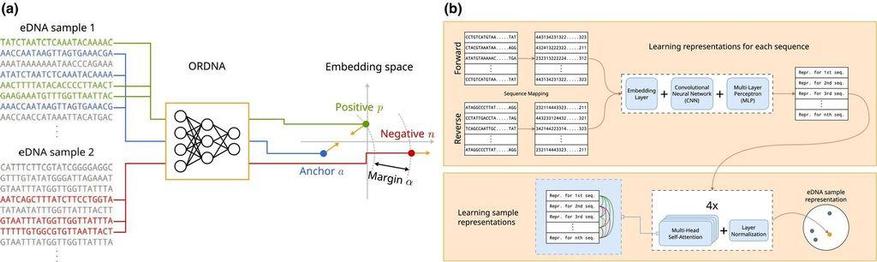
ORDNA: Deep-learning-based ordination for raw environmental DNA samples #eDNA #metabarcoding https://besjournals.onlinelibrary.wiley.com/doi/10.1111/2041-210X.70033
eDNAjoint: An R package for interpreting paired or semi-paired environmental DNA and traditional survey data in a Bayesian framework #eDNA #metabarcoding https://besjournals.onlinelibrary.wiley.com/doi/10.1111/2041-210X.70000
We are excited to share our new promo video for the #BIOLAWEB project!
With narration by Srđan Miletić, this video showcases all the activities realised during the BIOLAWEB project thanks to the incredible team, colleagues, and students.
Happy to announce a new paper of my 0hd student Paula Gauvin:
An annotated reference library for supporting DNA metabarcoding analysis of aquatic macroinvertebrates in French freshwater environments
https://mbmg.pensoft.net/article/137772/
#metabarcoding #eDNA #reference #library @pensoft @mbmg
Pipeline release! nf-core/ampliseq v2.13.0 - Ampliseq Version 2.13.0!
Please see the changelog: https://github.com/nf-core/ampliseq/releases/tag/2.13.0
Une offre de post-doc de 1 an renouvelable 1 an :
> Finaliser le développement d’une méthode d’identification moléculaire du phytoplancton utilisant le séquençage ADN haut-débit #metabarcoding
> CDD post-doc de 1 an, reconductible 1 an : démarrage à partir de mai 2025, Lieu : UMR Carrtel, Thonon https://fr-carrtel.lyon-grenoble.hub.inrae.fr/
> Projet pluriannuel #OFB et le projet européen #eDNAquaPlan
> candidature : frederic.rimet@inrae.fr
> plus de détails : https://filesender.renater.fr/?s=download&token=81a4c9dc-5e0f-49eb-a5bc-d83b53abb14d
and this paper also in MBMG erally nice : good work Tiina
https://mbmg.pensoft.net/article/130834/
Technology Readiness Level of biodiversity monitoring with molecular methods – where are we on the road to routine implementation?
by Laamanen et al.
#edna #methods #TRL #metabarcoding #standardisation
Researchers partner with #IndigenousKnowledge holders to protect threatened great desert skink Tjakuṟa https://phys.org/news/2025-01-partner-indigenous-knowledge-holders-threatened.html
#Metabarcoding clarifies the diet of the elusive and vulnerable Australian #tjakura (Great Desert #Skink, Liopholis kintorei) https://www.frontiersin.org/journals/ecology-and-evolution/articles/10.3389/fevo.2024.1354138/full
"Tjakuṟa is a large #lizard that lives in multi-tunneled communal burrows in #Australia... scientists rely on #Aṉangu Traditional Knowledge to locate Tjakuṟa burrows, estimate burrow occupancy, identify predator tracks"
https://mastodon.social/@aquaecomics/113837658626432987
aquaecomics - Happy to announce the program of AquaEcOmics meeting !!!
#omics #eDNA #metabarcoding #genomics #evian #nature #freshwater #marine
Access the program here:
https://aquaecomics.symposium.inrae.fr/program2
Really happy of our new paper:
Proficiency testing and cross-laboratory method comparison to support standardisation of diatom DNA metabarcoding for freshwater biomonitoring
By Valentin Vasselon, Sinzi Rivera and Benoît Paix et al.
#metabarcoding #eDNA #diatom #mbmg #science #nature #omics #calibration
https://mbmg.pensoft.net/article/133264/
Quick #introduction.
I am a #terrestrial #ecologist interested in #polar
#ecosystems. Currently #postdoc at #VUAmsterdam working on #Antarctic
#microbes
and #invertebrates
living in #mosses and #lichens.
When not doing #science, I take photos (ingeborgklarenberg.nl), and spend as much time as possible outside #running
, #climbing
, hiking and bird watching.
#eDNA #metabarcoding #nitrogenfixation #molecularecology
#symbiosis